Harnessing AI to Design Custom DNA for Targeted Liver Cancer Therapy: A Deep Dive into Carl Bisaga’s Exotic Model
Hello, fellow biotech enthusiasts! Today, let’s explore an exciting journey into the fusion of artificial intelligence and DNA design, narrated by Carl Bisaga, a fourth-year medical student from Chicago. Carl showcases his custom exotic model aimed at winning a cutting-edge DNA sequencing competition hosted by Exonic, a platform pioneering crowdsourced DNA design for precision medicine. Here’s a detailed walkthrough of his process, insights, and the revolutionary potential of AI in targeted chemotherapy drug development.
The Vision: Precision Medicine Meets AI-Driven DNA Design
Chemotherapy has long been a double-edged sword—effective in killing cancer cells but often wreaking havoc on healthy organs, causing severe side effects like hair loss and organ damage. Carl’s project is part of a larger mission to create DNA sequences that code for peptides specifically localized to the liver, thereby enabling drugs that target liver cancer cells with minimal off-target effects. Imagine a chemotherapy treatment that attacks only liver tumors without harming other tissues—this is the promise of precision medicine.
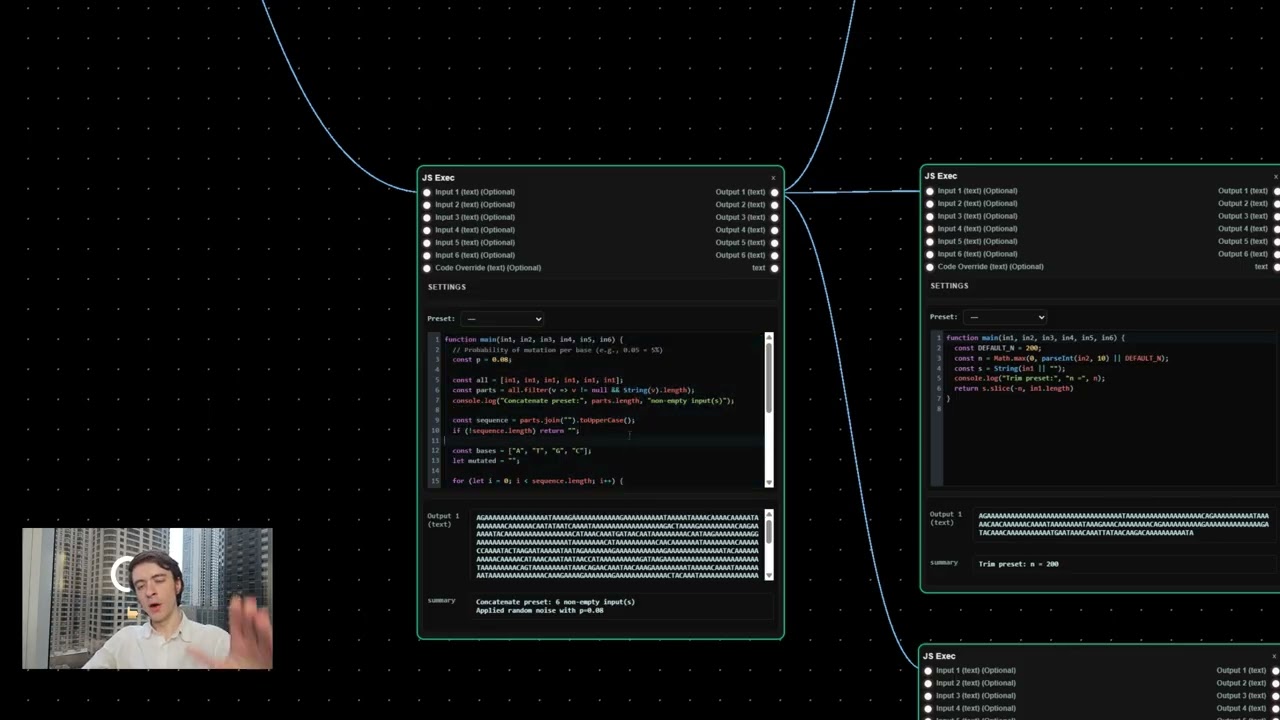
Phase 1: Generating Random DNA Sequences
The journey begins with creating random DNA sequences. These sequences initially hold little biological meaning—think of them as blank canvases consisting mostly of repeated adenine bases. Carl uses a DNA design panel within Exonic’s platform to generate these base sequences, which serve as the starting point for optimization.
Phase 2: Tweaking DNA with AI — The EVO2 Model
Random DNA isn’t enough. Carl integrates AI, specifically the EVO2 model, into his workflow to refine these sequences. The AI tidies up the DNA, making it more "organic" and increasing its likelihood of functioning well in wet lab experiments.
- Why longer sequences? Carl uses custom JavaScript to extend the DNA sequence to five times its original length before feeding it to EVO2. This provides the AI with richer context, enabling it to generate more meaningful and viable sequences.
- Parameters for AI tweaking: Temperature, Top K, and Top P control how aggressively EVO2 modifies the sequence. Adjusting these parameters allows balancing creativity and biological plausibility.
Phase 3: Testing and Validating DNA Sequences
No single metric can capture the effectiveness of a DNA sequence, so Carl evaluates multiple parameters to ensure the best candidates:
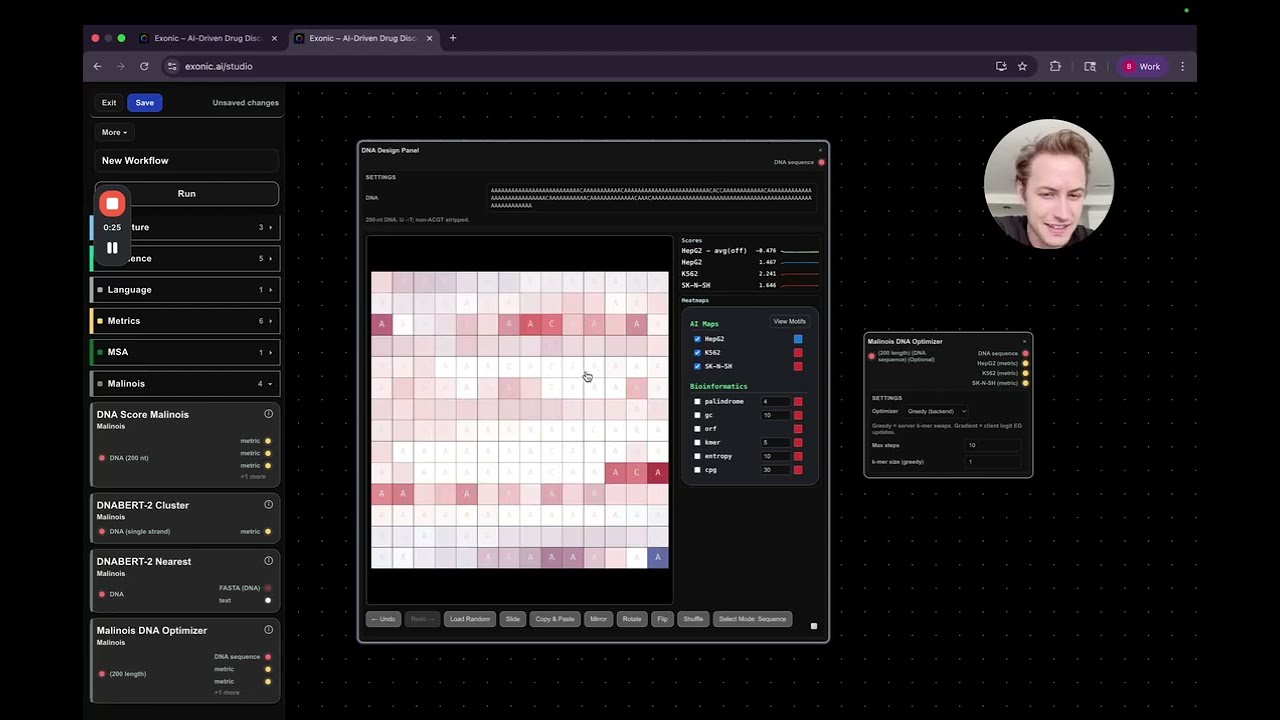
1. HEPG2 Score (Liver Localization)
- Derived from a melanoma DNA optimizer, the HEPG2 score estimates how well the peptide localizes to liver cells.
- More blue in the DNA visualizer indicates higher liver targeting, which is desirable.
- However, overfitting the HEPG2 metric (scores >6) may reduce overall effectiveness.
2. K562 Score (Blood Localization)
- Indicates how much the peptide localizes to blood cells.
- Lower scores are better since blood localization could cause systemic side effects.
3. SKNSH Score (Neuronal Localization)
- Reflects localization to neurons and the nervous system.
- Like K562, lower is preferred to avoid unwanted neurological side effects.
4. Perplexity Metric
- Measures how "natural" the AI believes the DNA sequence is.
- A score close to 1 indicates the sequence looks biologically plausible; higher scores suggest nonsense sequences.
5. DNA BERT 2 Nearest Metric
- Acts like a “Facebook for DNA,” comparing your sequence to a repository of 70,000 wet lab-tested DNA strands.
- Helps identify how similar your design is to known sequences, informing submission strategy.
6. Jasper Score
- Functions like LinkedIn for DNA, identifying transcription factors that bind to your DNA.
- Helps predict biological activity and tissue specificity by assessing which transcription factors interact with your sequence.
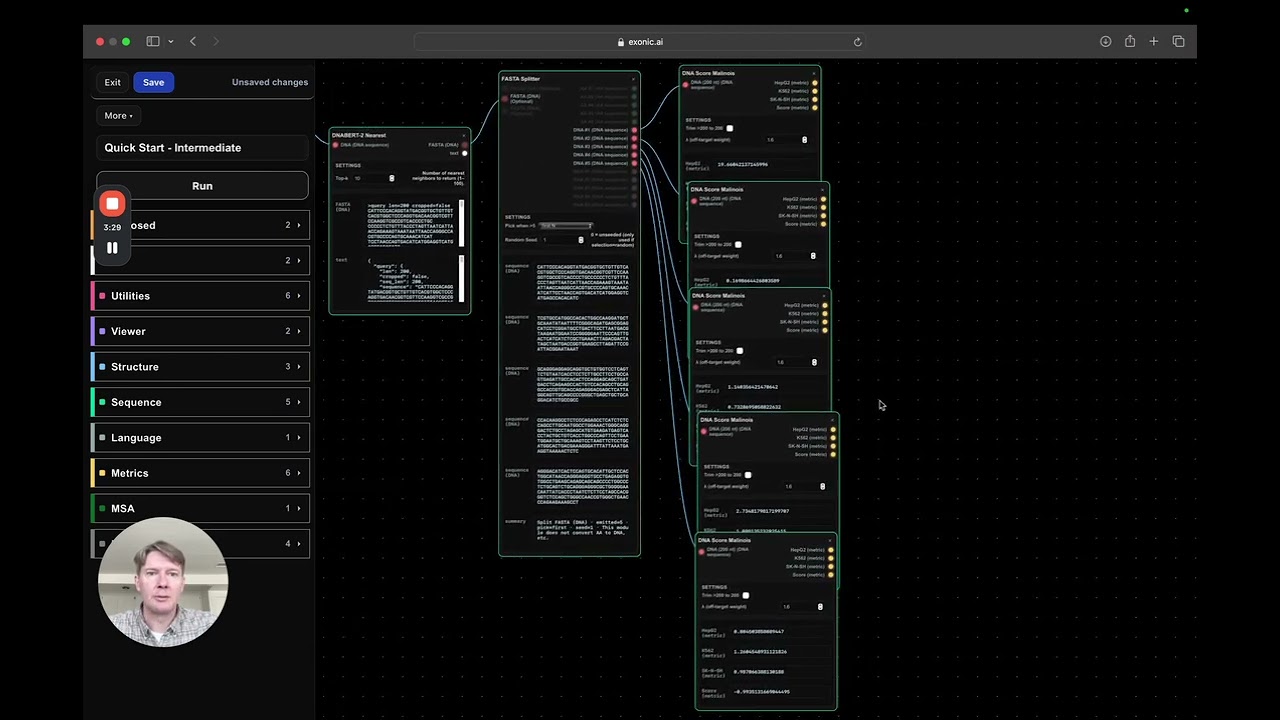
Putting It All Together: The Workflow in Action
Carl’s model pipeline looks like this:
- Generate random DNA → 2. Optimize with melanoma DNA optimizer → 3. Extend and refine with EVO2 AI → 4. Evaluate with multiple metrics (HEPG2, K562, SKNSH, Perplexity, DNA BERT, Jasper) → 5. Select top candidates for submission
This iterative process allows him to balance liver targeting while minimizing off-target effects, all while ensuring the DNA sequences remain biologically plausible.
Real-World Impact and Future Directions
Carl highlights the broader vision: leveraging AI’s ability to process and understand massive genetic sequences to revolutionize drug design. This approach could lead to highly specific chemotherapy drugs that drastically reduce side effects, improving patient quality of life.
He also emphasizes community involvement, encouraging others to experiment with parameters, explore different optimization nodes, and iterate continuously to find the best DNA sequences.
Final Thoughts
Carl’s model represents a powerful convergence of biology, medicine, and artificial intelligence. By thoughtfully combining random sequence generation, AI refinement, and multi-metric evaluation, he’s pushing the boundaries of what’s possible in DNA-based precision medicine.
As the competition unfolds, we look forward to seeing how these innovative designs perform in the wet lab and, ultimately, how they might transform liver cancer treatment.
About the Author
Carl Bisaga is a passionate fourth-year medical student and AI enthusiast based in Chicago. He actively contributes to the Exonic community, blending his medical knowledge with cutting-edge AI tools to advance the field of synthetic biology and precision medicine.
Good luck to all participants in the competition! Stay tuned for updates and breakthroughs in AI-driven DNA design.
If you’re interested in learning more about AI in biotechnology or want to try your hand at DNA sequence design, explore the Exonic platform and join the community pushing the frontiers of medicine.