[00:02] (2.40s)
Hello everybody. My name is Carl Bisaga.
[00:04] (4.72s)
Today I'm going to be showing you uh my
[00:07] (7.76s)
custom exotic model that I'm going to be
[00:10] (10.72s)
using to try to win this awesome
[00:13] (13.84s)
competition right here. Um I'm a fourth
[00:16] (16.96s)
year medical student here. I'm in
[00:18] (18.48s)
downtown Chicago today. A little bit of
[00:20] (20.56s)
foggy, but we're not going to let that
[00:22] (22.64s)
keep us down here. So, let's get right
[00:24] (24.80s)
into it. Um, I made this nice little uh
[00:28] (28.88s)
explainer, little PowerPoint screenshot
[00:31] (31.20s)
here for you today, um, to explain what
[00:33] (33.44s)
I'm going to do. So, here's the title.
[00:36] (36.24s)
We got how this model works. So, phase
[00:38] (38.32s)
one, first thing we're going to do is
[00:40] (40.00s)
create random DNA. Uh, this is
[00:42] (42.40s)
essentially DNA with very little meaning
[00:45] (45.20s)
attached to it, just something that we
[00:47] (47.60s)
uh, just sort of created on our own.
[00:49] (49.84s)
Then we move into phase two which is
[00:51] (51.68s)
tweaking the DNA with AI specifically
[00:54] (54.00s)
the EVO2 model which is conveniently
[00:56] (56.96s)
built into Exxonics platform. And then
[01:00] (60.40s)
we finish up with phase three which is
[01:02] (62.48s)
going to be testing the DNA uh according
[01:05] (65.12s)
to several metrics. No one metric is
[01:08] (68.00s)
perfect. So we're going to be looking
[01:10] (70.00s)
through several different uh metrics
[01:12] (72.32s)
including things like the um Malininoa
[01:15] (75.68s)
DNA optimizer metrics such as HEPG2A.
[01:18] (78.48s)
I'll explain this uh the details of that
[01:20] (80.96s)
later on. We're going to be looking at
[01:22] (82.08s)
Jasper scores. We're going to be looking
[01:23] (83.68s)
at um perplexity from the EVO 2. So,
[01:27] (87.36s)
let's dive right into it. So, I know
[01:29] (89.28s)
this is small resolution. We'll zoom in
[01:31] (91.04s)
a little bit here, but just broad
[01:33] (93.12s)
strokes. Here's what I'm working with
[01:34] (94.72s)
today. Here's my here's the grand
[01:36] (96.80s)
treasure map of my model. So, I went
[01:39] (99.44s)
ahead and organized it into sections
[01:42] (102.08s)
just as the uh as we covered in the plan
[01:44] (104.24s)
earlier. So this first rectangle here,
[01:46] (106.80s)
this is where we're creating the DNA. Uh
[01:49] (109.68s)
it's going to be very low meaning DNA.
[01:51] (111.76s)
It's just kind of random. Step two here,
[01:54] (114.40s)
this is going to be within this
[01:55] (115.76s)
rectangle right here. That is going to
[01:58] (118.32s)
be where we are engaging uh EVO 2. We're
[02:02] (122.24s)
feeding the DNA into the EVO 2 model and
[02:05] (125.44s)
it's going to tidy it up for us. It's
[02:08] (128.00s)
going to make it look organic so that it
[02:11] (131.04s)
has higher chance of success in the wet
[02:13] (133.20s)
lab in the uh final days of the
[02:15] (135.28s)
competition 47 days from now. And final
[02:20] (140.08s)
rectangle here that I'm uh showing with
[02:22] (142.96s)
my nice mechanical I got the little
[02:24] (144.80s)
cursor effects on my mouse here for you
[02:27] (147.92s)
guys today. Um this final part is the
[02:31] (151.60s)
part where we test the DNA sequence that
[02:34] (154.08s)
we've come up with because they can all
[02:36] (156.24s)
be winners. Let's be real.
[02:39] (159.12s)
So, and we only have three submissions
[02:41] (161.12s)
for the competition. So, we want to
[02:43] (163.20s)
choose our best three. So, let's dive
[02:45] (165.92s)
right into it. I'm going to go zoom in
[02:49] (169.12s)
Let's get this nice high resolution so
[02:51] (171.04s)
you can actually read what's going on
[02:52] (172.48s)
here. So, these little these little
[02:54] (174.88s)
panels, these are essentially the
[02:58] (178.00s)
building blocks of our machine. It's
[02:59] (179.68s)
almost like Legos. We get to plug and
[03:02] (182.08s)
play here. Uh you can find all types of
[03:05] (185.04s)
different uh nodes here in the sidebar.
[03:08] (188.72s)
I'm going to be covering a few of these
[03:10] (190.16s)
today, the highest yield ones for you,
[03:12] (192.48s)
but feel free to explore and uh take a
[03:15] (195.44s)
look at some of these other ones. Um
[03:18] (198.32s)
most crucial one we start off with here
[03:20] (200.24s)
is the DNA design panel. Essentially,
[03:24] (204.00s)
um looks like the just, you know, it
[03:26] (206.80s)
loaded in a sequence of DNA with uh all
[03:30] (210.88s)
adenosines here. is just straight
[03:32] (212.96s)
adenoses adenosines which it's probably
[03:36] (216.56s)
not biologically feasible. It basically
[03:38] (218.96s)
means nothing. So we can go and mess
[03:40] (220.48s)
around with it here. Uh what I like to
[03:42] (222.64s)
do is I like to drag this down a little
[03:45] (225.52s)
bit. And I just press load random a few
[03:47] (227.92s)
times. And let me tell you exactly what
[03:50] (230.48s)
we're looking at here. So why do these
[03:52] (232.96s)
colors change? Okay, we kind of have a
[03:54] (234.48s)
good one here. I'll explain why. So the
[03:56] (236.96s)
colors change according to um these
[04:00] (240.96s)
three metrics we have set out here. So
[04:03] (243.04s)
these three metrics they're der derived
[04:05] (245.12s)
from an earlier research paper. Um
[04:07] (247.60s)
essentially he g2 that is the good
[04:10] (250.56s)
metric. It outputs a number that we want
[04:13] (253.36s)
to go up. Uh so good will be blue here.
[04:16] (256.48s)
So the more blue we see in our little
[04:18] (258.72s)
DNA visualizer here, the better it is.
[04:21] (261.12s)
And let me explain why this is good. So
[04:24] (264.00s)
um I read through some of these papers
[04:25] (265.68s)
and essentially HEP G2. So the
[04:28] (268.72s)
overarching project we're working on
[04:30] (270.72s)
here at Exonic is well not me I'm part
[04:33] (273.28s)
of the community but um overall project
[04:36] (276.24s)
that this company's working on is
[04:39] (279.68s)
we are crowdsourcing DNA uh sequencing
[04:46] (286.08s)
because we feel that or because you know
[04:49] (289.12s)
blending the lines here a little bit. I
[04:50] (290.56s)
know Ben pretty well. He's a good friend
[04:51] (291.68s)
of mine. What can I say? So, I like to
[04:53] (293.60s)
say we um essentially we're creating DNA
[04:57] (297.20s)
strands. We're submitting them to this
[04:59] (299.44s)
competition and the DNA strands are
[05:02] (302.88s)
transcribed. Essentially, your the
[05:05] (305.60s)
proteins in your body read them almost
[05:07] (307.68s)
like a book. It's like a cookbook. It
[05:09] (309.84s)
tells them what protein to make, what
[05:11] (311.76s)
peptide to make. And this peptide is
[05:15] (315.52s)
essentially whatever peptide you make,
[05:18] (318.32s)
it actually doesn't quite matter. What
[05:20] (320.16s)
matters is where it localizes.
[05:22] (322.88s)
Um, we don't care too much about the
[05:24] (324.32s)
structure, but we do care about where
[05:25] (325.84s)
this protein is found. Because if we
[05:28] (328.24s)
happen to find the dream sort of that
[05:30] (330.88s)
we're working on this competition is if
[05:33] (333.04s)
we find a peptide that only is localized
[05:37] (337.68s)
to the liver when you when you uh give
[05:40] (340.32s)
it to a person as a uh as a medication.
[05:43] (343.36s)
If it only exists in the liver and it
[05:46] (346.40s)
doesn't exist anywhere else, that means
[05:48] (348.40s)
that we can rely upon this peptide, we
[05:53] (353.20s)
can take this peptide and attach to it a
[05:56] (356.72s)
huge um toxic chemical. And the reason
[06:01] (361.52s)
we want to do that is because there are
[06:02] (362.96s)
many cancers that occur in the liver.
[06:05] (365.20s)
And if we had a drug that only went to
[06:07] (367.28s)
those cancer cells in the liver and
[06:08] (368.88s)
destroyed them and didn't cause damage
[06:11] (371.12s)
anywhere else in the body, that would be
[06:12] (372.96s)
the definition of a perfect chemotherapy
[06:16] (376.16s)
drug. Um, that's also part of a
[06:18] (378.56s)
subbranch of what's called precision
[06:20] (380.16s)
medicine that's up and coming here in
[06:22] (382.16s)
the world. Um, the typical just a little
[06:25] (385.68s)
bit background in case you're not from
[06:27] (387.12s)
medical uh background. I always like to
[06:29] (389.12s)
give a little bit of context but uh the
[06:31] (391.28s)
chemotherapy drugs especially the
[06:33] (393.52s)
earlier uh generations of chemotherapy
[06:36] (396.24s)
drugs the problem is they they do
[06:39] (399.12s)
destroy the cancer but they cause
[06:42] (402.08s)
massive damage to every other organ
[06:44] (404.16s)
system in the body. That's why you
[06:45] (405.60s)
typically see uh you know these
[06:48] (408.08s)
unfortunate cancer patients they have to
[06:50] (410.08s)
shave their heads because the hair their
[06:53] (413.20s)
hair follicles are dying they're under
[06:55] (415.84s)
undergoing myopathy. Basically every
[06:57] (417.84s)
organ system is occurring some sort of
[06:59] (419.36s)
damage because this chemotherapy drug
[07:01] (421.68s)
just spreads throughout the body and
[07:02] (422.88s)
causes damage everywhere more so usually
[07:05] (425.44s)
in the cancer cells but still
[07:07] (427.60s)
systemically. So uh what essentially
[07:11] (431.28s)
what we're asking of of uh everyone in
[07:13] (433.28s)
the competition here is let's make
[07:15] (435.84s)
chemotherapy more specific. Let's make
[07:18] (438.32s)
it cause less damage. We don't want
[07:20] (440.24s)
people to have to shave their heads when
[07:22] (442.24s)
they have to uh do chemotherapy. we
[07:24] (444.56s)
wouldn't want them to have uh kidney
[07:26] (446.48s)
injuries and so forth, lung damage. Um
[07:29] (449.36s)
so that's a little bit about, you know,
[07:31] (451.52s)
that gives you so that gives you so much
[07:33] (453.20s)
more emotion that you can work with here
[07:34] (454.64s)
on the project. Uh aside like instead of
[07:37] (457.44s)
just looking at these little uh
[07:39] (459.36s)
anonymous names to the metrics. So um
[07:43] (463.04s)
the master plan here is HEB G2. HEP is
[07:46] (466.40s)
the Latin suffix for um sorry this Latin
[07:51] (471.20s)
prefix for liver. So, HEPG G2 means that
[07:56] (476.24s)
uh according to this model we have here,
[07:58] (478.32s)
this simple simple model, uh the more
[08:00] (480.88s)
blue we have in our square here, the
[08:03] (483.76s)
more it's going to localize in the
[08:05] (485.04s)
liver. And that is good. Um that's what
[08:08] (488.16s)
we want. K562 that corresponds to
[08:11] (491.76s)
localization in your blood. Uh just like
[08:14] (494.48s)
your serum, that means that it'll
[08:16] (496.64s)
generally hang out in your arteries,
[08:18] (498.72s)
your veins, your capillaries, and we
[08:21] (501.44s)
don't want that. um that is unwanted for
[08:24] (504.40s)
the purposes of chemotherapy or
[08:26] (506.56s)
amunotherapy. And for SKNSH that
[08:30] (510.24s)
corresponds to localizing the drug
[08:33] (513.20s)
localizing in the neurons. So
[08:35] (515.84s)
essentially your nervous system and
[08:38] (518.08s)
mostly your brain. Your brain is the
[08:39] (519.76s)
largest mass of that system. So that
[08:42] (522.40s)
means that these two metrics we actually
[08:44] (524.16s)
want them to be low. So that means we
[08:46] (526.16s)
don't want a lot a lot of red in our
[08:48] (528.56s)
visualizer here. So, um, we can just go
[08:53] (533.28s)
ahead keep on randomizing this just to
[08:55] (535.28s)
get a good framework to start off of
[08:57] (537.12s)
here. Sometimes it can be a little
[08:58] (538.32s)
difficult. So, I'm going to compromise.
[09:00] (540.16s)
We'll get a little bit of purple. You
[09:01] (541.44s)
know, sometimes life you got to
[09:02] (542.96s)
compromise a little bit. [laughter]
[09:05] (545.68s)
This is a fantastic start though. So, we
[09:07] (547.44s)
have our DNA sequence here. This is the
[09:09] (549.20s)
same DNA sequence sequence we see in our
[09:11] (551.84s)
image because it's linked to the
[09:14] (554.08s)
Melanino DNA optimizer. it will
[09:16] (556.80s)
specifically so this tool it
[09:19] (559.60s)
specifically increases it aims to
[09:22] (562.08s)
increase the HEPG2 metric and it seeks
[09:25] (565.04s)
to decrease the localization in the
[09:28] (568.48s)
blood the K5 and the neurons the SK SK
[09:32] (572.96s)
um and the way it does this is it's a
[09:34] (574.64s)
very simple simple um
[09:38] (578.32s)
simple al excuse me algorithm um it is
[09:43] (583.12s)
not nearly as complex as the EVO 2 model
[09:46] (586.00s)
which is why I tend to put it in the
[09:48] (588.08s)
beginning and let the AI model clean up
[09:50] (590.32s)
afterwards. The parameters we can change
[09:53] (593.28s)
here max steps. So this is where the
[09:55] (595.84s)
wisdom of crowds come in. Um I
[09:58] (598.64s)
essentially am play putting these
[10:00] (600.16s)
there's no correct answer for these
[10:02] (602.08s)
numbers putting into uh to these
[10:04] (604.40s)
parameters here. If you started off with
[10:06] (606.16s)
a much better sequence perhaps you want
[10:09] (609.04s)
to go lighter on these. Essentially,
[10:11] (611.04s)
this means that I'm allowing the uh
[10:13] (613.68s)
optimizer to make 40 changes
[10:17] (617.36s)
uh to my DNA. And each of those changes
[10:19] (619.92s)
will be three base pairs or letters in
[10:23] (623.60s)
length um at a time. You could change
[10:26] (626.00s)
this. If you really want to be drastic,
[10:27] (627.52s)
you change it to four and so forth.
[10:29] (629.52s)
We'll stick with three here. That's a
[10:30] (630.88s)
good starting point. Um once I click run
[10:33] (633.84s)
here, then this sequence will output
[10:36] (636.96s)
into here. So this is not the sequence
[10:39] (639.12s)
that we see on the image here. That'll
[10:40] (640.96s)
only appear once we run this whole
[10:42] (642.80s)
thing. So I'm going to leave that like
[10:44] (644.08s)
it is for now.
[10:46] (646.56s)
Okay. So that's it for phase one where
[10:48] (648.32s)
we created the random DNA. We tweaked it
[10:50] (650.80s)
a little bit with some simple uh
[10:52] (652.88s)
algorithms here. That word is giving me
[10:56] (656.56s)
a hard time today. [laughter]
[10:58] (658.64s)
Um and the next step of this uh just as
[11:02] (662.00s)
a quick reminder here we are tweaking
[11:03] (663.60s)
the DNA with AI. So the way that I like
[11:06] (666.32s)
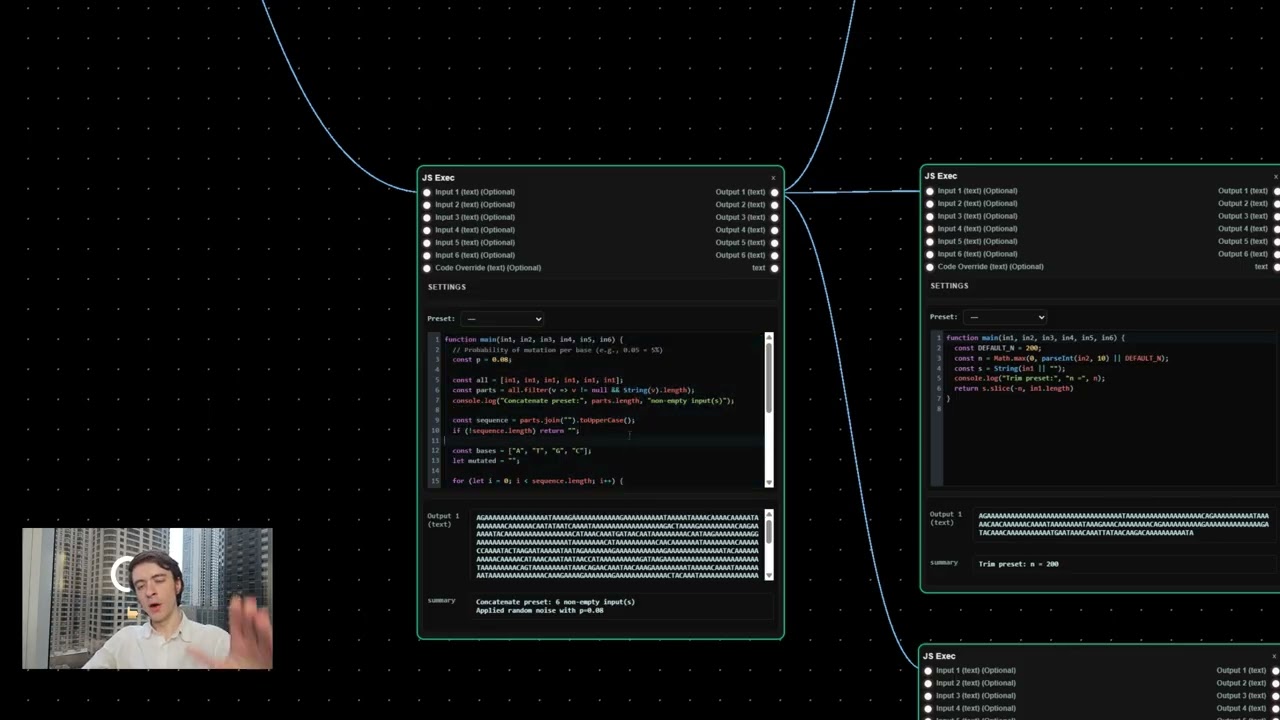
to do that is I've written a little code
[11:09] (669.12s)
here in JavaScript. Essentially it takes
[11:11] (671.04s)
that uh randomized DNA here as an input
[11:14] (674.72s)
and using JavaScript you know oh man
[11:17] (677.76s)
code I have to code really Carl like
[11:20] (680.00s)
you're really going to do that to me I
[11:21] (681.28s)
have to spend four hours of my Sunday
[11:23] (683.20s)
evening. No. So I'm going to explain
[11:25] (685.20s)
this to you. You can copy paste this
[11:26] (686.56s)
code. Essentially this code takes the
[11:28] (688.80s)
DNA that we have and it extends it to uh
[11:32] (692.40s)
five times its length. As you can see
[11:34] (694.56s)
here, this is a much longer series of
[11:36] (696.48s)
base pairs. Um, and the reason why we
[11:39] (699.12s)
want that is because we are going to
[11:41] (701.44s)
feed that longer sequence into the EVO2
[11:44] (704.24s)
model. And why that's valuable is
[11:45] (705.92s)
because essentially if imagine we gave
[11:49] (709.92s)
uh let's move it to the absolute
[11:51] (711.52s)
extremes here. If we gave the EVO2 model
[11:54] (714.40s)
one single base pair just an adenosine
[11:57] (717.20s)
and we told it okay generate a sequence
[12:00] (720.00s)
of 200 base pairs that is going to be
[12:02] (722.80s)
valuable here uh for our chem or for our
[12:05] (725.84s)
wet lab it's going to be total nonsense
[12:07] (727.84s)
because it only has a single data point
[12:10] (730.08s)
and on the same uh on the most on the
[12:12] (732.64s)
other opposite side of the spectrum
[12:14] (734.24s)
imagine we gave it a million base pairs
[12:15] (735.84s)
and we told it to generate something
[12:17] (737.92s)
that is very useful it'll probably
[12:21] (741.04s)
generate something more useful in the
[12:24] (744.64s)
longer sequence. So essentially this is
[12:26] (746.80s)
taking advantage of the way the AI model
[12:29] (749.92s)
works. It likes more data. It likes a
[12:32] (752.00s)
longer prompt to give you a better
[12:33] (753.68s)
answer. And we've we're inputting
[12:36] (756.88s)
this,200 base pair sequence into it and
[12:40] (760.16s)
it will go ahead and um change it
[12:43] (763.20s)
according to these parameters.
[12:44] (764.48s)
Essentially um this is how long we want
[12:46] (766.96s)
our output to be. So you can still you
[12:49] (769.20s)
still have to output a 200 base pair uh
[12:52] (772.16s)
DNA sequence. That's what you're going
[12:53] (773.52s)
to submit in the competition here. Uh so
[12:55] (775.60s)
you're not going to change this, but you
[12:57] (777.04s)
are going to uh play around with these
[13:00] (780.00s)
uh few parameters. Temperature is
[13:02] (782.56s)
essentially how aggressive you want the
[13:04] (784.32s)
AI model to be in your changes. Top K
[13:06] (786.88s)
and top P. I'm not going to cover too
[13:08] (788.32s)
much today. Seems like a great job for
[13:11] (791.04s)
GPT to be to be quite honest. Um, and
[13:16] (796.56s)
once it does that, it's going to output
[13:18] (798.88s)
its final DNA sequence. That's going to
[13:21] (801.76s)
be the last thing that we change here.
[13:23] (803.52s)
And it's going to put it into the final
[13:25] (805.60s)
section, phase three, which is testing
[13:27] (807.36s)
our DNA. As we can see here, uh, we So,
[13:31] (811.20s)
it's like I said earlier, we're going
[13:32] (812.72s)
through a few different tests because no
[13:34] (814.48s)
one test is perfect. It's the same way
[13:37] (817.44s)
that you can't pick a boyfriend off of
[13:40] (820.08s)
one date. You got to, you know, you got
[13:42] (822.16s)
to go visit his parents. You got to go
[13:44] (824.00s)
ask him about his favorite movies. You
[13:46] (826.00s)
got to annoy him a little bit. See how
[13:47] (827.28s)
he responds, right? So, so this is the
[13:50] (830.16s)
same thing with our DNA here. Um
[13:55] (835.92s)
let's go ahead. Yeah. So let's cover
[13:57] (837.92s)
this. So the HEPG2, this is the metric I
[14:00] (840.72s)
talked about earlier. Um there's some
[14:03] (843.68s)
some data that s suggests that
[14:07] (847.28s)
uh HEBG2 can be overfitted if the number
[14:11] (851.36s)
goes too high. that number is six. So if
[14:13] (853.92s)
it's over six, there is a slight danger
[14:16] (856.72s)
of overfitting your DNA to um this
[14:20] (860.72s)
metric which is a you know no metric is
[14:23] (863.60s)
perfect, right? So um it might not
[14:26] (866.80s)
necessarily be the best strategy in the
[14:29] (869.36s)
competition to uh make this number very
[14:32] (872.80s)
large such as in the 20s, 30s, 40s which
[14:35] (875.28s)
I've seen in the competition which I've
[14:36] (876.80s)
also submitted. Um so that is one risk
[14:40] (880.24s)
with this metric. these K562, you want
[14:43] (883.12s)
this to be below one as a starting
[14:44] (884.96s)
point. Ideally, the closer to zero it
[14:47] (887.36s)
is, the better it is because that means
[14:48] (888.96s)
that, you know, way on down the line, 10
[14:51] (891.28s)
years from now, the future patients
[14:52] (892.96s)
getting perhaps um your drug that you
[14:56] (896.56s)
help design. You know, the lower this
[14:59] (899.36s)
is, the less side effects they're going
[15:00] (900.80s)
to have in their different organ
[15:01] (901.92s)
systems. You know, if you're curious,
[15:03] (903.68s)
there actually are other metrics that
[15:05] (905.36s)
judge how much the drug or the sequence
[15:09] (909.92s)
localizes in the lungs, for example.
[15:11] (911.92s)
There's other uh other se, you know,
[15:15] (915.12s)
every organ is at play here. So, it's
[15:17] (917.60s)
not just going to be these few metrics.
[15:19] (919.12s)
This is just to start off here. So, it's
[15:20] (920.56s)
a a future jumping off point if you
[15:22] (922.96s)
really wanted to nail down your DNA
[15:24] (924.56s)
sequence. Um, but and then finally, this
[15:27] (927.68s)
is just a composite score of the f of
[15:29] (929.52s)
the three up here. I'm not quite sure
[15:32] (932.32s)
how much it weights each individual
[15:34] (934.08s)
metric.
[15:36] (936.40s)
Um, as for DNA BERT 2 nearest, so this
[15:39] (939.84s)
is a neat little tool that essentially
[15:43] (943.68s)
it is almost like a Facebook of DNA
[15:46] (946.24s)
sequences. It takes your DNA sequence,
[15:49] (949.28s)
which is almost like a name, you type in
[15:50] (950.88s)
on Facebook, and it returns the
[15:54] (954.32s)
uh top closest
[15:57] (957.36s)
um hits that are match your DNA, the
[16:00] (960.64s)
closest,
[16:02] (962.48s)
and it gives you a similarity score. And
[16:04] (964.88s)
the nice thing is it's searching through
[16:07] (967.28s)
a repository of 70,000
[16:10] (970.56s)
uh wet lab already tested DNA sequences.
[16:13] (973.84s)
So if your sequence is very similar to a
[16:16] (976.96s)
sequence that has already been tested in
[16:18] (978.64s)
the lab and already performs very well
[16:20] (980.40s)
or maybe perhaps not very well that can
[16:23] (983.20s)
give you a better decision on whether or
[16:24] (984.72s)
not to submit this DNA to the wet lab.
[16:27] (987.28s)
Um the scores you're looking at here
[16:28] (988.88s)
similarity goes from zero to one. One is
[16:31] (991.52s)
more similar uh to the listed sequence
[16:35] (995.52s)
from the repository that's already been
[16:37] (997.44s)
tested and distance is the inverse of
[16:39] (999.92s)
similarity. Um, and it gives you quite a
[16:43] (1003.20s)
few results here. So, you can walk
[16:46] (1006.48s)
through this just as you like. So,
[16:47] (1007.84s)
that's quick explainer on DNA BERT. That
[16:50] (1010.32s)
was our second little metric or a little
[16:52] (1012.80s)
test that we gave to our DNA sequence.
[16:55] (1015.28s)
And then the final one I'm going to
[16:56] (1016.40s)
cover here today. There are a few others
[16:58] (1018.48s)
uh down here. These are not too
[16:59] (1019.84s)
important. This is more so fine-tuning
[17:02] (1022.16s)
uh that'll perhaps be important if
[17:04] (1024.32s)
you're really trying to lock in your DNA
[17:06] (1026.64s)
choice here. The final test here is the
[17:09] (1029.20s)
Jasper score. And this is important
[17:12] (1032.00s)
because again it's it if that was
[17:15] (1035.12s)
Facebook the if the uh DNA BERT was like
[17:17] (1037.76s)
Facebook Jasper is almost like LinkedIn.
[17:20] (1040.48s)
It searches through it doesn't search
[17:23] (1043.12s)
through DNA sequences but it does search
[17:25] (1045.76s)
through transcription factors that have
[17:28] (1048.96s)
already been well researched on the uh
[17:32] (1052.08s)
and uploaded to the internet essentially
[17:34] (1054.00s)
in an open- source repository. And this
[17:36] (1056.64s)
score tells you how active this specific
[17:39] (1059.28s)
transcription factor is. Um so for
[17:43] (1063.04s)
example
[17:44] (1064.56s)
um this DNA sequence that was submitted
[17:48] (1068.64s)
to Jasper, it it searched the DNA
[17:52] (1072.48s)
sequence and it looked at what
[17:53] (1073.76s)
transcription factors bind to this DNA
[17:56] (1076.16s)
sequence. And the top one top hit here
[17:59] (1079.60s)
is ZNF-558. And we can go ahead and look
[18:02] (1082.88s)
up more info about what ZNF558 does. And
[18:06] (1086.24s)
that is very useful because we can
[18:08] (1088.00s)
theorize, you know, if ZNF-558 is only
[18:11] (1091.52s)
active in the brain and this is your top
[18:14] (1094.40s)
transcription factor, probably not a
[18:16] (1096.72s)
good sign for your DNA sequence. But if
[18:18] (1098.64s)
it only is active in the liver, then we
[18:21] (1101.68s)
might be uh on to something with our DNA
[18:24] (1104.48s)
sequence. uh the score above two I
[18:27] (1107.68s)
believe that means that trips
[18:29] (1109.12s)
transcription factor is really going to
[18:30] (1110.88s)
enjoy bind buying binding to your DNA
[18:33] (1113.84s)
and that is going to be particularly
[18:36] (1116.08s)
more active uh with your sequence and
[18:38] (1118.72s)
there are many perhaps too many
[18:41] (1121.52s)
transcription factors returned here
[18:43] (1123.36s)
there's hundreds here so you can really
[18:46] (1126.00s)
get into the nitty nitty-gritty of the
[18:47] (1127.92s)
macro of the microbiology here so that
[18:51] (1131.28s)
is it for the quick run through here
[18:52] (1132.96s)
let's go ahead and pause the video I'm
[18:54] (1134.96s)
going to go ahead and run my machine and
[18:56] (1136.72s)
see what comes out here.
[18:59] (1139.36s)
All right, everyone. So, our model
[19:01] (1141.92s)
finished running. It took about a minute
[19:03] (1143.52s)
and a half here. Very fast. So, I'll
[19:06] (1146.08s)
walk you through what it did step by
[19:07] (1147.60s)
step here. Once again, it took this DNA
[19:10] (1150.56s)
sequence. This is unchanged from how we
[19:12] (1152.48s)
had it. Now, these metrics are finally
[19:15] (1155.44s)
accurate. They correspond to our current
[19:19] (1159.12s)
uh DNA sequence that we generated here
[19:21] (1161.36s)
randomly. As you can see, terrible,
[19:23] (1163.52s)
terrible scores.
[19:25] (1165.76s)
uh it's going all over the blood, it's
[19:27] (1167.44s)
going all over the neurons, it's causing
[19:28] (1168.96s)
a lot of damage, the patient's not
[19:30] (1170.56s)
happy, they're ticked off. So, we went
[19:33] (1173.36s)
ahead and optimized it with the Melanino
[19:35] (1175.28s)
DNA optimizer,
[19:37] (1177.36s)
and it made some changes to our DNA
[19:39] (1179.76s)
sequence, improved our score. HEPG2
[19:42] (1182.64s)
actually went down a little bit, but
[19:44] (1184.48s)
thankfully the K5 and the SK uh were cut
[19:48] (1188.32s)
down a little bit. particularly the K5
[19:51] (1191.68s)
dropped down by two points and the SK
[19:54] (1194.56s)
still not the greatest here. Um, ideally
[19:57] (1197.36s)
want that to be under 0.6 I would say
[20:00] (1200.40s)
probably even lower than that, but
[20:01] (1201.84s)
that's a good starting point here just
[20:03] (1203.68s)
to display what this model is up to. We
[20:06] (1206.16s)
extended it. We ran it through the AI
[20:09] (1209.20s)
model, the EVO 2 revolutionary tech here
[20:11] (1211.92s)
from 2024.
[20:14] (1214.24s)
And uh one thing I didn't touch on
[20:16] (1216.16s)
earlier but uh is important to know is
[20:19] (1219.28s)
uh this perplexity metric is important
[20:21] (1221.12s)
because it it is the AI telling you how
[20:24] (1224.80s)
organic your DNA sequence looks. Um for
[20:28] (1228.64s)
instance, it's the equivalent of someone
[20:32] (1232.48s)
coming up to you and saying a random
[20:34] (1234.96s)
series of words and this score
[20:37] (1237.68s)
essentially would be how do you think
[20:40] (1240.96s)
this person is a human or not? Because
[20:42] (1242.88s)
there are certain things that you know
[20:45] (1245.12s)
you can write down certain sequences of
[20:46] (1246.88s)
DNA that are total nonsense chemically
[20:49] (1249.04s)
and biologically and the AI the nice
[20:51] (1251.76s)
thing is the AI will recognize that and
[20:54] (1254.08s)
when it is total nonsense it will give
[20:56] (1256.88s)
you a very high score. it'll be over 10.
[20:59] (1259.52s)
Generally, the closer it is to one means
[21:02] (1262.24s)
that your DNA sequence actually
[21:04] (1264.96s)
corresponds to the the the AI model
[21:08] (1268.00s)
believes it is something that could be
[21:09] (1269.60s)
seen in nature, which is very good. Um,
[21:13] (1273.92s)
and then finally for our so we ran it
[21:16] (1276.72s)
through the model and so these are the
[21:18] (1278.24s)
final scores we're looking at. These are
[21:20] (1280.00s)
the the finishing scores. HEP G2
[21:22] (1282.80s)
actually went down a little bit. Uh, but
[21:25] (1285.60s)
the K5, this is actually the lowest K5
[21:27] (1287.52s)
I've ever seen. Wow. 0.03. That's very
[21:29] (1289.76s)
low. Very good. Um, you know, it's
[21:32] (1292.40s)
probably lower actually on the
[21:33] (1293.52s)
competition. I haven't looked at it too
[21:34] (1294.72s)
much. Um, the SK just under that 0.6
[21:38] (1298.88s)
mark. So, it seemed to emphasize
[21:41] (1301.52s)
lowering the K5 and the SK more instead
[21:44] (1304.08s)
of increasing the HEP G2. Uh that could
[21:46] (1306.88s)
be something that you could one strategy
[21:50] (1310.08s)
to mitigate that would be to put another
[21:51] (1311.92s)
Malininoa DNA optimizer here and
[21:54] (1314.08s)
optimize it once again after running it
[21:56] (1316.16s)
through the EVO 2 model. You could
[21:58] (1318.56s)
essentially just run this in a loop over
[22:01] (1321.04s)
and over until you reach some sort of
[22:03] (1323.04s)
local maximum or perhaps the global
[22:05] (1325.76s)
maximum as we're hoping here and uh
[22:08] (1328.48s)
perhaps potentially get a fantastic uh
[22:10] (1330.80s)
DNA sequence to put in the competition.
[22:13] (1333.44s)
And as for DNA Bur, as we can see here,
[22:17] (1337.44s)
we look at our, this is our Facebook
[22:20] (1340.72s)
test, right? So, we looking at what DNA
[22:24] (1344.40s)
sequences are closest to the ones we've
[22:26] (1346.32s)
input into the model. And looks like
[22:28] (1348.88s)
we're very, very different from the
[22:32] (1352.08s)
closest cousin we could find on our
[22:34] (1354.64s)
repository of 70,000 sequences here. So,
[22:37] (1357.92s)
our sequence is actually very different,
[22:40] (1360.72s)
which could be good or bad depending on
[22:43] (1363.36s)
what your strategy here is in the
[22:44] (1364.96s)
competition. And then finally looking at
[22:47] (1367.28s)
Jasper,
[22:48] (1368.88s)
if we really had good scores on the
[22:50] (1370.96s)
HEPG2 and so forth and the uh DNA Bert
[22:55] (1375.36s)
was also looking good strategically,
[22:58] (1378.24s)
then we would move into the Jasper
[23:00] (1380.00s)
scores and really dive down deep into
[23:01] (1381.60s)
the weeds here and figure out, okay,
[23:03] (1383.84s)
what is PX PBX1 do inside of the body?
[23:08] (1388.16s)
um according to the latest research, how
[23:10] (1390.56s)
do we theorize that this will help our
[23:14] (1394.16s)
DNA sequence localize to the liver um to
[23:17] (1397.04s)
the HEPG2 cells and kill those pesky
[23:20] (1400.40s)
cancer cells. So that is it for it for
[23:23] (1403.28s)
me today everyone. Um I'm glad you
[23:25] (1405.68s)
really excited about this competition.
[23:27] (1407.44s)
Feel like we can make a huge impact
[23:28] (1408.72s)
here. This is all about you know the
[23:30] (1410.32s)
using the dream of AI. We're using
[23:32] (1412.48s)
cutting edge technology. The value of a
[23:36] (1416.48s)
machine that can understand a sequence
[23:38] (1418.32s)
that is millions of base pairs long that
[23:40] (1420.48s)
a human could never understand is uh
[23:43] (1423.12s)
revolutionary. And I'm really excited
[23:44] (1424.88s)
about this project. Really happy I could
[23:46] (1426.48s)
walk you through it today. And for me
[23:49] (1429.36s)
today, that is it. Um I'm going to run
[23:52] (1432.40s)
out downtown today and go get a haircut.
[23:55] (1435.60s)
So good luck in the competition
[23:57] (1437.68s)
everyone. I'll see you in the
[23:58] (1438.80s)
leaderboards. Thanks.
 YouTube Deep Summary
YouTube Deep Summary Extract content that makes a tangible impact on your life